Biomimetic systems for actin-based motility
 The movement of cells and
membrane shape changes is largely powered by the formation and
disassembly of actin filament networks adjacent to cell membrane. One
of the challenges in understanding cell motility is to dissect the
elementary biochemical and biophysical steps that link actin
polymerization to mechanical force generation. Research using in vitro
bio-mimetic systems has led to significant progress in understanding
actin based motility, by reducing the enormous complexity of motile
cells.
We use a system of artificial phospholipid vesicles coated with an
actin nucleating factor. Once added to a solution containing actin and
other necessary proteins, they self-assemble into motile vesicles
propelled by polymerizing actin "comet" tails. The actin filaments
in the comet tail are tightly cross-linked into an elastic gel. The
polymerization forces cause significant deformation of the moving
vesicles. A measure of the spatial and temporal distribution of forces
can be obtained by analyzing the curvature of the vesicle contour, a
purely geometric quantity. A major advantage of this in vitro approach
is that both biochemical and physical parameters can be controlled
precisely to obtain quantitative information about the mechanism of
force generation.
Actin filaments are semi-flexible polymers which are crosslinked
together to form a network. Changing the composition of the
crosslinking proteins will alter the structure of the actin gel as
well as its mechanical properties. One outstanding question is how the
forces generated depend on the structure and mechanical properties of
actin networks. We will use tools from soft condensed matter physics
- both experimental and theoretical - to study the
mechanical behavior of reconstituted cytoskeletal networks.
The movement of cells and
membrane shape changes is largely powered by the formation and
disassembly of actin filament networks adjacent to cell membrane. One
of the challenges in understanding cell motility is to dissect the
elementary biochemical and biophysical steps that link actin
polymerization to mechanical force generation. Research using in vitro
bio-mimetic systems has led to significant progress in understanding
actin based motility, by reducing the enormous complexity of motile
cells.
We use a system of artificial phospholipid vesicles coated with an
actin nucleating factor. Once added to a solution containing actin and
other necessary proteins, they self-assemble into motile vesicles
propelled by polymerizing actin "comet" tails. The actin filaments
in the comet tail are tightly cross-linked into an elastic gel. The
polymerization forces cause significant deformation of the moving
vesicles. A measure of the spatial and temporal distribution of forces
can be obtained by analyzing the curvature of the vesicle contour, a
purely geometric quantity. A major advantage of this in vitro approach
is that both biochemical and physical parameters can be controlled
precisely to obtain quantitative information about the mechanism of
force generation.
Actin filaments are semi-flexible polymers which are crosslinked
together to form a network. Changing the composition of the
crosslinking proteins will alter the structure of the actin gel as
well as its mechanical properties. One outstanding question is how the
forces generated depend on the structure and mechanical properties of
actin networks. We will use tools from soft condensed matter physics
- both experimental and theoretical - to study the
mechanical behavior of reconstituted cytoskeletal networks.
Cell adhesion and migration
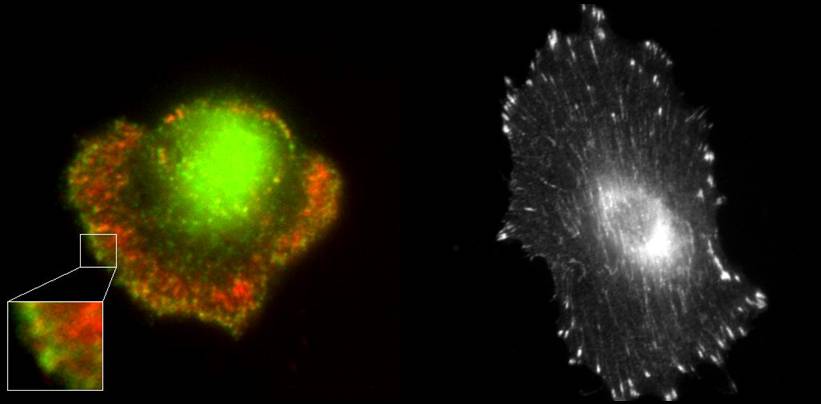
Cell adhesion and migration is essential for embryonic development, wound healing and proper functioning of the immune system. Cell movement requires the coordination of various modules such as attachment to a substrate via adhesion bonds, protrusion of the membrane at the leading edge of the cell using actin polymerization and retraction of the rear of the cell facilitated by the contraction of actin myosin networks. Impairment of these functions can cause invasive behavior of cells leading to cancer metastasis. Before cells can move, they need to establish an attachment to the substrate. When cells are plated on a substrate, they initiate attachments on the order of milliseconds to seconds. This is followed by a rapid increase in attached area by the formation of new adhesions which takes tens of minutes - in other words the cell spreads over the substrate. The final stage over a timescale of hours involves the formation of focal contacts and focal adhesions. These are clusters of adhesion molecules and cytoskeletal components which act as mechano-sensory sites that allow the cell to sense and generate forces required for movement and change in shape. We would like to understand the mechanics of the process of cell spreading and movement. How do the physical properties of the cellular cytoskeleton, cell membrane and the extracellular environment regulate the dynamics of these processes? We will use a combination of techniques such as high resolution fluorescence imaging of GFP-tagged proteins in live cells, high-speed imaging, micro-fabrication to perturb the cell substrate interface and optical trapping to measure forces exerted by the cytoskeleton and membrane. At a theoretical level, statistical mechanics and continuum mechanics approaches will be used to model these phenomena.
Morphogenetic rearrangements
Cells in tissues need to move in a coordinated manner to create coherent structures. However, at the macroscopic scale, it is possible to ignore the molecular details of individual cells and their constituents and focus on certain generic physical properties. Surface forces arising from differences in adhesion drive rearrangement resulting in segregation of different cell types (sorting) or spreading of one cell type over another (engulfment). The collective spreading of cells and tissues occurs during morphogenesis and wound healing. Patterning of tissues during development, such as in the retina of Drosophila, is in part determined by surface mechanics. Spreading of a cell aggregate on an adhesive substrate resembles the wetting of a surface by a liquid droplet.
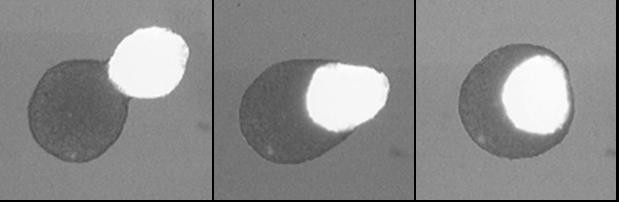 |
Engulfment of embryonic heart tissue (bright sphere) by neural retinal tissue (dark sphere). Scale bar is 300 microns. |
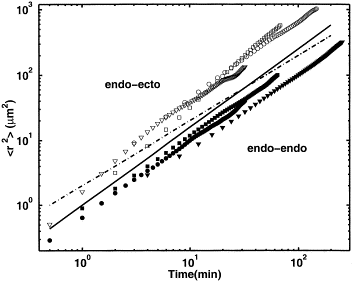 |
Cell motion within cellular aggregates consists of both random and coherent components. We used confocal microscopy to study the center of mass displacements and deformations of single endodermal Hydra cells in two kinds of cellular aggregates, ectodermal and endodermal. In both aggregates, cells perform a persistent random walk, with the diffusion constant smaller in the more cohesive endodermal aggregate. |
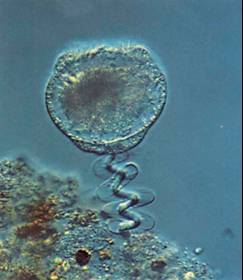
Vorticella contraction
Vorticella convallaria is one of the fastest and most powerful biological springs. The cell body is attached to a substrate by a slender stalk containing a rod-like polymeric structure - the spasmoneme. Helical coiling of the stalk results from rapid contraction of the spasmoneme using a calcium dependent mechanism. Binding of calcium by the Ca-binding proteins called spasmins is believed to be the principal source for the contraction. In the absence of calcium, the spasmin filaments are negatively charged and in an extended state due to electrostatic repulsion. Calcium binding neutra lizes the charges and leads to an entropic collapse of the spasmoneme.We have conducted high-speed imaging experiments to study the contraction dynamics. These include measuring the contraction velocity and force as a function of increasing viscous load to obtain force-velocity curves. We have also used beads as markers on the stalk to obtain important information about the signal propagation mechanism that leads to contraction. We find that the spasmoneme behaves as a self-reinforcing spring and that the contraction starts at the cell body and proceeds down the stalk.
 |
Time series of Vorticella contraction (time shown in milliseconds). The scale bar is 35 microns. The initially straight stalk bends and coils into a helix, starting from the region near the cell body and moving down towards the base of the stalk with continuously changing helical pitch. The cell body moves without any observable rotation until the end of the contraction. After the stalk becomes fully coiled, the zooid starts rotating in a clockwise direction. |
Using optical traps to study intracellular membranes
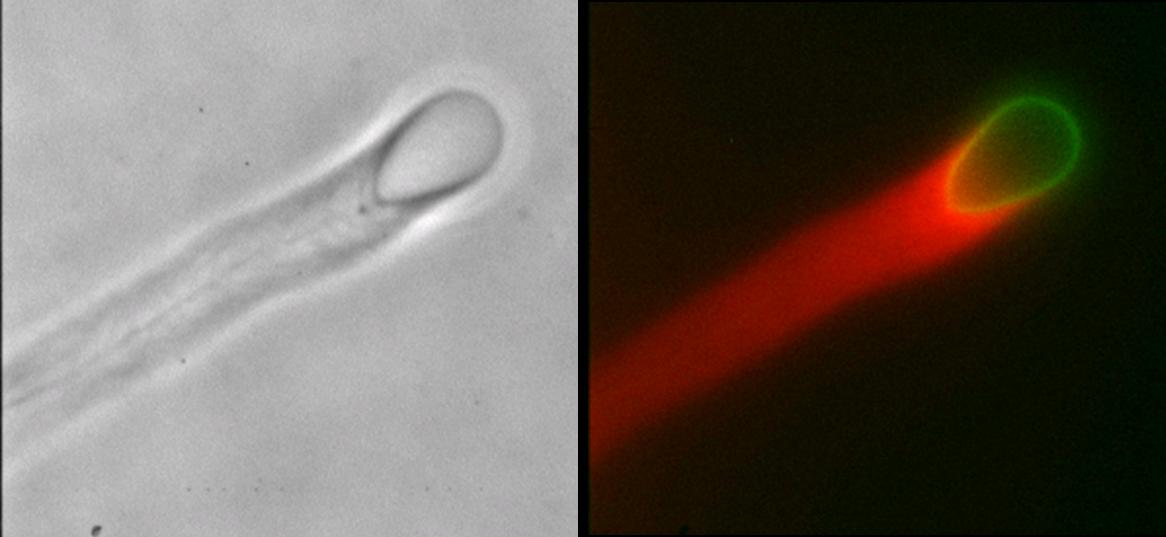
The endoplasmic reticulum (ER) and Golgi have robust bidirectional traffic between them and yet form distinct membrane compartments. Membrane tubules are pulled from large aggregates of ER or Golgi by microtubule motors to form ER tubulovesicular networks or Golgi tubules both in vivo and in vitro. The physical properties of membranes are critical for membrane traffic and organelle morphology. For example, tension applied to membranes can create tethers, drive membrane flow, and set the diameter of the tubules. Here, we formed ER and Golgi membrane networks in vitro and used optical tweezers to measure directly, for the first time, the membrane tensions of these organelles to clarify the possible role of tension in membrane flow. We report that higher forces are needed to form tethers from ER (18.6 �� 2.8 pN) than from Golgi (11.4 �� 1.4 pN) membrane tubules in vitro. Since ER tubules are smaller in diameter than Golgi tubules, it follows that Golgi networks have a lower tension than ER. The higher tension of the ER could be an explanation of how Golgi tubules can be rapidly drawn into the ER by tension-driven flow after fusion, as is observed in vivo.
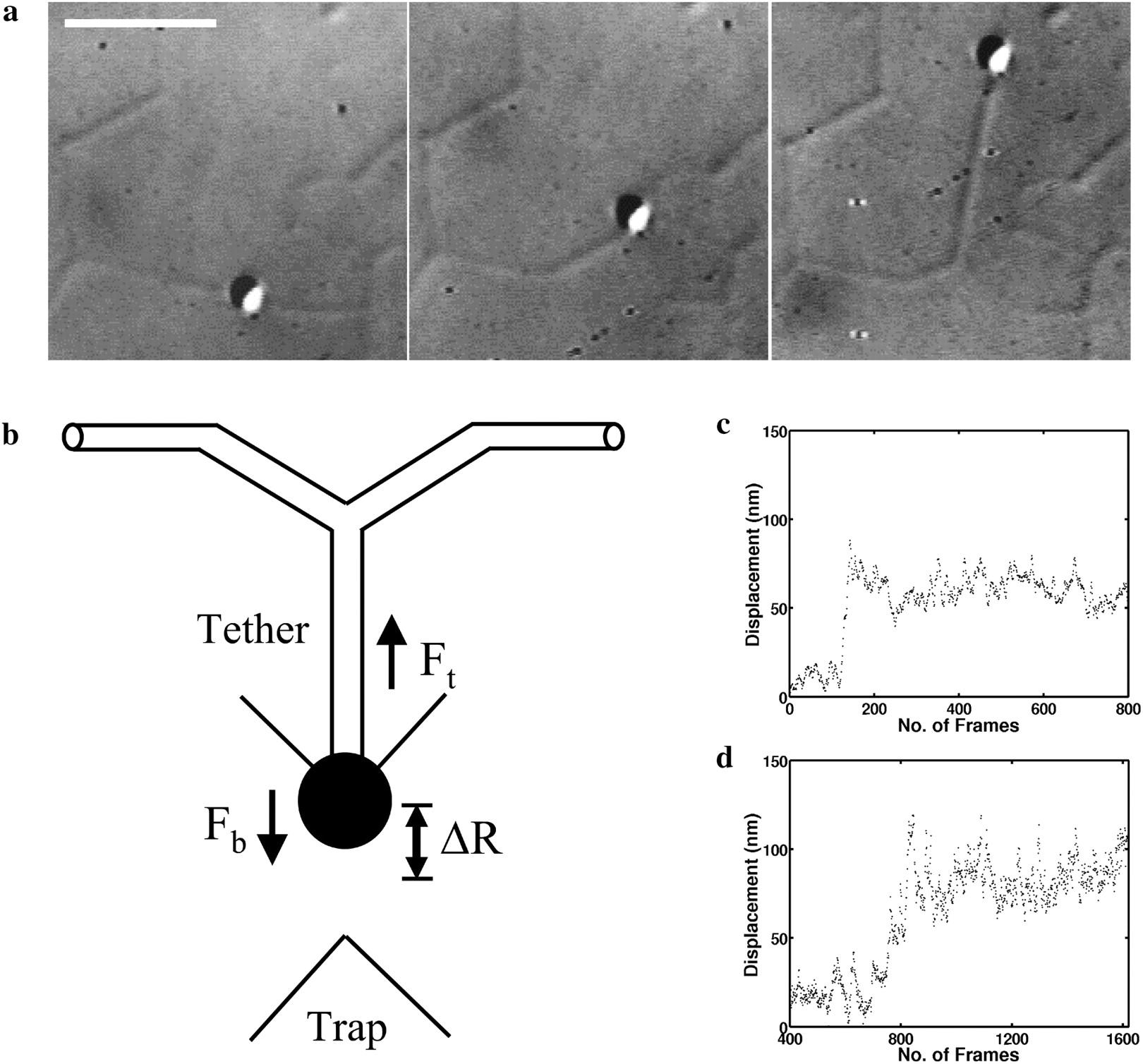 |
Force measurement using optical tweezers. (a) DIC image of a typical tether-pull sequence. The bead is held in the optical trap and pulled orthogonal to the membrane tubule.(b) Schematic of a tether-pull showing the displacement of a trapped bead from the trap center. The tether force Ft balances the trap force Fb pulling the bead toward the trap center. (c and d) Typical curves showing the displacement of a trapped bead after pulling a membrane tether from a branch for networks from the Golgi (c) and the ER (d). On the x axis, 30 frames correspond to 1 s. |
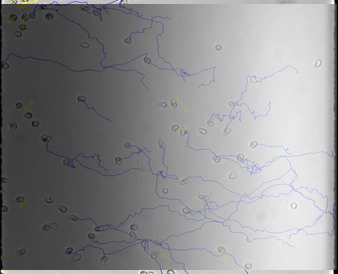
Chemotaxis in controlled gradients
The mechanism of neutrophil chemotaxis in response to a chemokine is well defined and represents a central paradigm by which the migration of this cell type towards a site of injury is understood. Studies of neutrophil chemotactic responses to the chemokine and known chemoattractant IL-8, have shown that gradient sensing and the directional decisions of neutrophils are dependent on the polarization and signaling of G-protein coupled receptors. Studies of neutrophil and D. discoideum motility have also revealed that an extracellular gradient of a chemokinetic agent results in intracytoplasmic accumulation of proteins differentially at the leading and trailing edges. In order to study cell motility in external chemical gradients, we have developed quantitative measures to characterize the movement of human neutrophils in precise chemoattractant gradients prepared using microfluidic devices (collaboration with M. Poznansky, Harvard Medical School).
 |
|